The EpiNext™ Post-Bisulfite DNA Library Preparation Kit is a complete set of optimized reagents to prepare a DNA library -- after successful bisulfite conversion -- for various Illumina platform-based bisulfite sequencing (bisulfite-seq) assays, such as whole genome bisulfite sequencing (WGBS), oxidative bisulfite sequencing (oxBs-seq), reduced representation bisulfite sequencing (RRBS), and other bisulfite-based next generation sequencing applications. The optimized protocol and components of the kit allow both non-barcoded (singleplexed) and barcoded (multiplexed) DNA libraries to be quickly constructed using sub-nanogram input concentrations of DNA since the DNA is first bisulfite-converted and then used for library preparation. The kit has the following advantages and features: - Allows bisulfite-converted DNA to be used directly for ligation, thereby eliminating the possibility of breaking adapter-ligated fragments, which can often occur in currently used WGBS and RRBS methods.
- Fast 5-hour procedure, from input starting material to library amplification.
- Gel-free size ion/purification saves time and prevents handling errors, as well as loss of valuable samples.
- High sensitivty and efficiency -- direct ligation of adapter to bisulfite-converted DNA fragments reduces loss of fragments and ion bias, which enables pre-bisulfite input DNA to be as low as 1 ng. The kit can be used for both non-barcoded (singleplexed) and barcoded (multiplexed) DNA library preparation.
- Comprehensive set of components to accommodate each step of DNA library preparation -- ligation, clean-up, size ion, and library amplification -- for convenience, consistency, and reliability.
- Ultra HiFi amplification enables achievement of reproducibly high yields of DNA libraries with minimal sequence bias and low error rates.
Background Information
Several methods, including whole genome bisulfite sequencing (WGBS) and reduced representation bisulfite sequencing (RRBS), are currently used for genome-wide DNA methlyation analysis. These methods convert unmethylated cytosines to uracil while 5-methylcytosines remain unchanged by the bisulfite treatment. This allows epigenetic differences to become genetic differences, which can be subsequently detected via sequencing at the single-based resolution level and on a genome-wide scale. However, practical use with such current methods are not ideal as they (1) need large amounts of DNA (>1 µg) as input material, which is difficult to prepare from limited biological samples such as tumor biopsy, early embyros, embyronic tissues, and circulating DNA; (2) require DNA to first be sheared and then ligated to adapters, followed by bisulfite conversion (post-ligation bisulfite conversion), which causes substantial amounts of DNA fragments ontained in the adapter-DNA fragment constructs to be broken and thereby forms mono-tagged templates that become removed during library enrichment; and (3) are time-consuming (2 days). The EpiNext Post-Bisulfite DNA Library Preparation Kit is designed to overcome these weaknesses. Principle & Procedure
With this kit, bisulfite-treated DNA (which is in single-stranded form), is converted to double-stranded DNA and directly used for ligation with BisDNA-specific adapters which are necessary for amplification and sequencing. The fragments are then size ed and purified with MQ beads, which allows for quick and precise size ions of DNA. Size-ed DNA fragments are then amplified with a high-fidelity PCR Mix, ensuring maximum yields from minimum amounts of starting material and providing a highly accurate amplification of library DNA with low error rates and minimal bias. Starting Materials
Input starting material must be bisulfite-treated DNA generated from various input DNA amounts of 1 ng to 1 µg. For optimal preparation, the input DNA amount for the bisulfite conversion process should be 100 ng to 200 ng so that sufficient bisulfite-treated DNA can be yielded. | 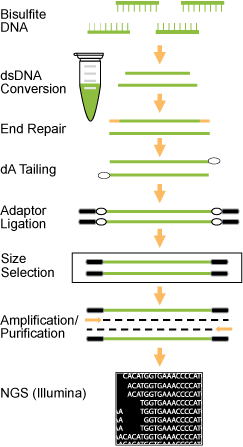
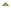 Fig. 1. Schematic procedure for the EpiNext™ Post-Bisulfite DNA Library Preparation Kit. Fig. 1. Schematic procedure for the EpiNext™ Post-Bisulfite DNA Library Preparation Kit.
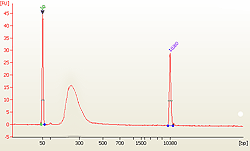
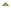 Fig. 2. Size distribution of library fragments as demonstrated by a post-bisulfite DNA library constructed using the EpiNext™ Post-Bisulfite DNA Library Preparation Kit from 10 ng of input DNA. Fig. 2. Size distribution of library fragments as demonstrated by a post-bisulfite DNA library constructed using the EpiNext™ Post-Bisulfite DNA Library Preparation Kit from 10 ng of input DNA.

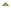 Fig. 3. The EpiNext™ Post-Bisulfite DNA Library Preparation Kit was used to prepare a library for Methyl-Seq with an Illumina HiSeq 2500. Read alignment data at 19 kb and base pair resolution in chromosome 7 shows CpG methylation differences for treated and control samples. Control samples are unmethylated (blue) while treated samples are methylated (red). Fig. 3. The EpiNext™ Post-Bisulfite DNA Library Preparation Kit was used to prepare a library for Methyl-Seq with an Illumina HiSeq 2500. Read alignment data at 19 kb and base pair resolution in chromosome 7 shows CpG methylation differences for treated and control samples. Control samples are unmethylated (blue) while treated samples are methylated (red).
|