·产品描述
组蛋白修饰是一种表观遗传修饰.翻译后的组蛋白修饰包括组蛋白乙酰转移酶介导的组蛋白赖氨酸残基乙酰化,组蛋白去乙酰化酶介导的去乙酰化,组蛋白甲基转移酶介导的赖氨酸和精氨酸的甲基化,组蛋白去甲基化酶介导的去甲基化,组蛋白磷酸化酶可以特定丝氨酸磷酸化,其他的修饰包括泛素化,SUMO化和聚ADP-核酸单位.除了DNA甲基化之外,组蛋白乙酰化和甲基化是最重要的表观遗传标记.总的说来,H3-K4, H3-K36或是H3-K79的三甲基化会导致一个开放的染色质构型,因而是常染色质的代表.常染色质以很高的组蛋白乙酰化为标志,而此特点是组蛋白乙酰化酶介导的.赖氨酸残疾可以被单、双或是三甲基化,每一种都可以不同程度上调节染色质结构和转录.还有其他的组蛋白修饰,比如磷酸化,这些修饰可以有不同的组合.以上这些组合形成了一个“组蛋白密码”,可以通过不同的细胞因素进行解读.
EpiQuik组蛋白H3修饰定量试剂盒(比色法)是一组完全的、优化的试剂组合,可以同时定量H3上面21个修饰方式,是一个简单的Elisa检测方法.
·产品特点
1.同时检测21种不同的组蛋白H3修饰方式
2.反应快速,在2.5h内完成
3.创新的比色法让实验者摆脱了同位素,电泳,色谱或是其他昂贵的仪器
4.高灵敏度是够检测限达到0.5ng/孔,可检测20ng-500ng/孔的组蛋白提取物
5可以检测每一种H3修饰
6.含有H3组蛋白标准品,可以对各种样品进行标准化
7.实验灵活,可以进行单个修饰的处理,也可以进行21个修饰的处理
8.额外的2条8孔板可以帮助进行上样量的优化,从而使检测结果落在检测限以内(20 ng to 500 ng/孔)
9方案简单,可靠,稳定
| Input Type: | Histone Extracts |
| Research Area: | Histone Methylation |
| Target Application: | Amount Quantitation |
| Vessel Format: | 96-Well Plate |
| 100% Guarantee: | 6 months |
The EpiQuik™ Histone H3 Modification Multiplex Assay Kit (Colorimetric) is a complete set of optimized reagents to detect and quantify up to twenty-one (21) modified histone H3 patterns simultaneously in a simple, ELISA-like format with use of a standard microplate reader. The kit has the following advantages and features:
Background Information
Histone modifications have been defined as epigenetic modifiers. Post-translational modifications (PTMs) of histones include the acetylation of specific lysine residues by histone acetyltransferases (HATs), deacetylation by histone deacetylase (HDACs), the methylation of lysine and arginine residues by histone methytransferases (HMTs), the demethylation of lysine residues by histone demethylases (HDMTs), and the phosphorylation of specific serine groups by histone kinases (HKs). Additional histone modifications include the attachment of ubiquitin (Ub), small ubiquitin-like modifiers (SUMOs), and poly ADP-ribose (PAR) units. Next to DNA methylation, histone acetylation and histone methylation are the most well characterized epigenetic marks. Generally, tri-methylation at H3-K4, H3-K36, or H3-K79 results in an open chromatin configuration and is therefore characteristic of euchromatin. Euchromatin is also characterized by a high level of histone acetylation, which is mediated by histone acetyltransferases. Lysine residues can be mono-, di-, or tri-methylated, each of which can differentially regulate chromatin structure and transcription. Along with other histone modifications such as phosphorylation, this enormous variation leads to a multiplicity of possible combinations of different modifications. This may constitute a “histone code”, which can be read and interpreted by different cellular factors.
Principle & Procedure
In an assay with this kit, each histone H3 modified at specific sites will be captured by an antibody that is coated on the strip wells and specifically targets the appropriate histone modification pattern. The captured histone modified at specific sites will be detected with a detection antibody, followed by a color development reagent. The ratio of modified histone is proportional to the intensity of absorbance measured by a microplate reader at a wavelength of 450 nm.
Starting Materials
Input materials can be histone extracts or purified histone H3 proteins. The amount of histone extracts for each assay can be 20 ng to 500 ng with an optimal range of 50 ng to 100 ng depending on the purity of histone extracts. The amount of purified histone H3 proteins for each assay can be 1 ng to 25 ng with an optimal range of 4 ng to 5 ng.
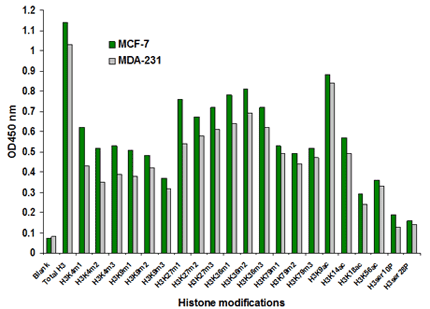
 Fig. 3. Histone extracts were prepared from MCF-7 and MDA-231 cells using the EpiQuik™ Total Histone Extraction Kit (Cat. No. OP-0006) and multiple histone H3 modifications were screened and measured using the EpiQuik™ Histone H3 Modification Multiplex Assay Kit (Colorimetric). 100 ng of total histone proteins were used.
Fig. 3. Histone extracts were prepared from MCF-7 and MDA-231 cells using the EpiQuik™ Total Histone Extraction Kit (Cat. No. OP-0006) and multiple histone H3 modifications were screened and measured using the EpiQuik™ Histone H3 Modification Multiplex Assay Kit (Colorimetric). 100 ng of total histone proteins were used.
Product Citations
Shandilya R. et. al. et. al. (January 2021). Immuno-cytometric detection of circulating cell free methylated DNA, post-translationally modified histones and micro RNAs using semi-conducting nanocrystals Talanta. 222
Hirano K et. al. (July 2020). Indonesian Ginger (Bangle) Extract Promotes Neurogenesis of Human Neural Stem Cells through WNT Pathway Activation. Int J Mol Sci. 21(13)
Nair A.M et. al. et. al. (March 2020). Polymer coated Fibre Optic Sensor as a Process Analytical Tool for Biopharmaceutical Impurity Detection IEEE.
Fuentes-Mattei et. al. (January 2020). miR-543 regulates the epigenetic landscape of myelofibrosis by targeting TET1 and TET2 JCI Insight. 5(1)
Bhat et. al. (December 2019). The Dopamine Receptor Antagonist TFP Prevents Phenotype Conversion and Improves Survival in Mouse Models of Glioblastoma bioRxiv.
Al Sayed R et. al. (December 2019). A 2x folic acid treatment affects epigenetics and dendritic spine densities in SHSY5Y cells. Biochem Biophys Rep. 20:100681.
Roy S et. al. (November 2019). Phloroglucinol Treatment Induces Transgenerational Epigenetic Inherited Resistance Against <i>Vibrio</i> Infections and Thermal Stress in a Brine Shrimp (<i>Artemia franciscana</i>) Model. Front Immunol. 10:2745.
Gobert AP et. al. (October 2019). Bacterial Pathogens Hijack the Innate Immune Response by Activation of the Reverse Transsulfuration Pathway. MBio. 10(5)
Gambacurta A et. al. (June 2019). Human osteogenic differentiation in Space: proteomic and epigenetic clues to better understand osteoporosis. Sci Rep. 9(1):8343.
Bayo J et. al. (March 2019). A comprehensive study of epigenetic alterations in hepatocellular carcinoma identifies potential therapeutic targets. J Hepatol.
Peixoto P et. al. (February 2019). EMT is associated with an epigenetic signature of ECM remodeling genes. Cell Death Dis. 10(3):205.
Mukherjee K et. al. (February 2019). Epigenetic mechanisms mediate the experimental evolution of resistance against parasitic fungi in the greater wax moth Galleria mellonella. Sci Rep. 9(1):1626.
Zhou R et. al. (November 2018). Characterization of H3 methylation in regulating oocyte development in cyprinid fish. Sci China Life Sci.
Yang L et. al. (May 2018). Differential gene regulatory plasticity between upper and lower layer cortical excitatory neurons. Mol Cell Neurosci.
Parira T et. al. (September 2017). Novel detection of post-translational modifications in human monocyte-derived dendritic cells after chronic alcohol exposure: Role of inflammation regulator H4K12ac. Sci Rep. 7(1):11236.
Seth C et. al. (December 2016). Long-Lasting WNT-TCF Response Blocking and Epigenetic Modifying Activities of Withanolide F in Human Cancer Cells. PLoS One. 11(12):e0168170.
Bandyopadhaya A et. al. (October 2016). A quorum-sensing signal promotes host tolerance training through HDAC1-mediated epigenetic reprogramming. Nat Microbiol. 1:16174.
Lee HO et. al. (July 2016). Lack of global epigenetic methylation defects in CBS deficient mice. J Inherit Metab Dis.
Rao A et. al. Switch to autophagy the key mechanism for trabecular meshwork death in severe glaucoma. Research Square.