·产品描述
DNA中的5-甲基胞嘧啶主要与基因的表达抑制有关,但是同时5-mC也被发现在很多的RNA分子中出现,如tRNAs, rRNAs, mRNAs和ncRNAs(非编码RNA).至少10275个5-mC候选位点在mRNA和ncRNA上被发现,占转录组总胞嘧啶的10.6%.5-mC在某些种类的ncRNA中含量丰富,但在mRNA中相对较少.但是,主要的候选位点(83%)在mRNA中.在这些转录产物中,5-mC似乎在蛋白编码序列中较少,而在5’和3’的非翻译区较多.两种不同的甲基转移酶,NSUN2和DNMT2可以催化5-mC在真核RNA的修饰.最近的数据显示RNA胞嘧啶的甲基化影响很多生物过程的调控,比如RNA稳定性和mRNA转录.此外,穹窿体RNA的5-mC的缺失会导致与Argonaute蛋白相关的小RNA片段的异常处理. 因此,穹窿体 ncRNA的处理过程的被破坏 会导致一些与NSUN2缺陷相关的人类疾病. RNA的重亚硫酸盐转化,配合后续的RT-PCR,克隆和测序产生可靠地RNA胞嘧啶甲基化程度的信息.
Methylamp RNA重亚硫酸盐转化试剂盒 是一组完全的用于重亚硫酸盐转化RNA样本的试剂组合.通过Methylamp RNA重亚硫酸盐转化试剂盒制备的RNA适合下游RNA甲基化分析,包括甲基化特异性RT-PCR,MS-HRM和重亚硫酸盐RNA测序(如焦磷酸测序和deep测序)
·产品特点
1.快速的反应方案使实验在3h内完成
2.完全把未甲基化的RNA胞嘧啶转化成尿嘧啶(大于99.9%)
3.强大的保护功能使得RNA不会降解,减少了超过90%的RNA损失量
4.含有内参引物,可以测试重亚硫酸盐转化的程度
5.少量RNA样本即可检测,每个反应RNA量少至5ng
6.简单,可靠及稳定的检测条件
| Input Type: | RNA |
| Research Area: | RNA Methylation |
| Target Application: | Sample Modification |
| Vessel Format: | Columns/Tubes |
| 100% Guarantee: | 6 months |
The Methylamp™ RNA Bisulfite Conversion Kit is a complete set of optimized reagents required for fast bisulfite conversion on an RNA sample. The converted RNA obtained with the Methylamp RNA Bisulfite Conversion Kit is suitable for various downstream RNA methylation analyses including methylation-specific RT-PCR, MS-HRM, and bisulfite RNA-sequencing (e.g., pyrosequencing and deep-sequencing). The kit has the following features and advantages:
Background Information
5-methylcytosine (5-mC) in DNA been well studied and is generally associated with repression of gene expression, but it has also been observed that in humans, 5-mC occurs in various RNA molecules including tRNAs, rRNAs, mRNAs, and non-coding RNAs (ncRNAs). At least 10,275 5-mC candidate sites were discovered in mRNAs and ncRNAs, which covered 10.6% of the total cytosine residues in the transcriptome. 5-mC seems to be enriched in some classes of ncRNA, but relatively depleted in mRNAs. However, the majority (83%) of their candidate sites were found in mRNAs. Within these transcripts, 5-mC appears to be depleted within protein-coding sequences but enriched in 5’ and 3’ UTRs. Two different methyltransferases, NSUN2 and DNMT2 are known to catalyze the 5-mC modification in eukaryotic RNAs. Recent data strongly suggest that RNA cytosine methylation affects the regulation of various biological processes such as RNA stability and mRNA translation. Furthermore, loss of 5-mC in vault RNAs causes aberrant processing into Argonaute-associated small RNA fragments that can function as microRNAs. Thus, impaired processing of vault ncRNA may contribute to the etiology of human disorders related to NSUN2-deficiency human disorders. Bisulfite conversion of RNA, followed by RT-PCR amplification, cloning, and sequencing yields reliable information about RNA cytosine methylation states. By treating RNA with bisulfite, cytosine residues are deaminated to uracil while leaving 5-methylcytosine intact:
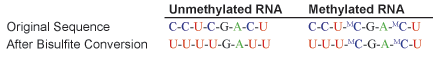
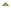 Bisulfite conversion of RNA, followed by RT-PCR amplification, cloning, and sequencing, yields reliable information about RNA cytosine methylation states. By treating RNA with bisulfite, cytosine residues are deaminated to uracil while leaving 5-methylcytosine intact.
Bisulfite conversion of RNA, followed by RT-PCR amplification, cloning, and sequencing, yields reliable information about RNA cytosine methylation states. By treating RNA with bisulfite, cytosine residues are deaminated to uracil while leaving 5-methylcytosine intact.
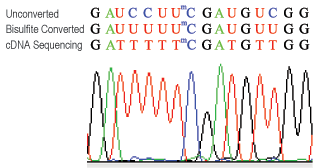
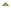 RNA bisulfite sequencing analysis. RNA isolated from MCF-7 cells is subject to bisulfite treatment, cDNA synthesis and sequencing after PCR amplification using 28S ribosomal RNA primers specific to bisulfite converted RNA. Unmethylated cytosines (#4, #5, #14) are converted to uracil and detected as thymine while methylated cytosine (#8) remains the same.
RNA bisulfite sequencing analysis. RNA isolated from MCF-7 cells is subject to bisulfite treatment, cDNA synthesis and sequencing after PCR amplification using 28S ribosomal RNA primers specific to bisulfite converted RNA. Unmethylated cytosines (#4, #5, #14) are converted to uracil and detected as thymine while methylated cytosine (#8) remains the same.
Principle & Procedure
The Methylamp RNA Bisulfite Conversion Kit contains all reagents required for fast and efficient bisulfite conversion on an RNA sample. A unique conversion mix solution contains powerful RNA protection reagents to prevent chemical and thermophilic degradation, thus leading to the accelerated conversion of all cytosines to uracil with negligible methylcytosine deamination. The non-toxic RNA capture solution enables RNA to tightly bind to the column filter, so that converted RNA cleaning can be carried out on the column to effectively remove residual bisulfite and salts.
Starting Materials
RNA isolated from various tissue or cell samples can be used as starting material. The amount of RNA for each bisulfite reaction can be 5 ng to 1 µg. For an optimal reaction, the input RNA amount should be 200 ng to 500 ng. When using the Methylamp RNA Bisulfite Conversion Kit for methylation-specific RT-PCR with very small amounts of input RNA (<10 ng), the number of PCR cycles should be greater than 45. The yield of RNA purified after bisulfite conversion depends on the amount of input RNA, nature of RNA, and source of the starting material.
Product Citations
Henry BA et. al. (April 2020). 5-Methylcytosine Modification of an Epstein-Barr Virus Noncoding RNA Decreases its Stability. RNA.
Xu X et. al. (October 2019). Advances in methods and software for RNA cytosine methylation analysis. Genomics.
Janin M et. al. (August 2019). Epigenetic loss of RNA-methyltransferase NSUN5 in glioma targets ribosomes to drive a stress adaptive translational program. Acta Neuropathol.